Opening a File
There are a variety of different file formats which can represent molecular data. PorphyStruct can handle the following files:
- .cif (Crystallographic Information File from CCDC Database or generated via ShelXL, OLEX, ...)
- .mol2 (TRIPOS Mol2 Files)
- .pdb (Protein Data Bank files, can contain proteins)
- .xyz (Simple Cartesian Coordinates, often used in computational chemistry. Bonds are generated on the fly)
- .mol (MOL File Standard by MDL Information Services, used e.g by ChemSpider)
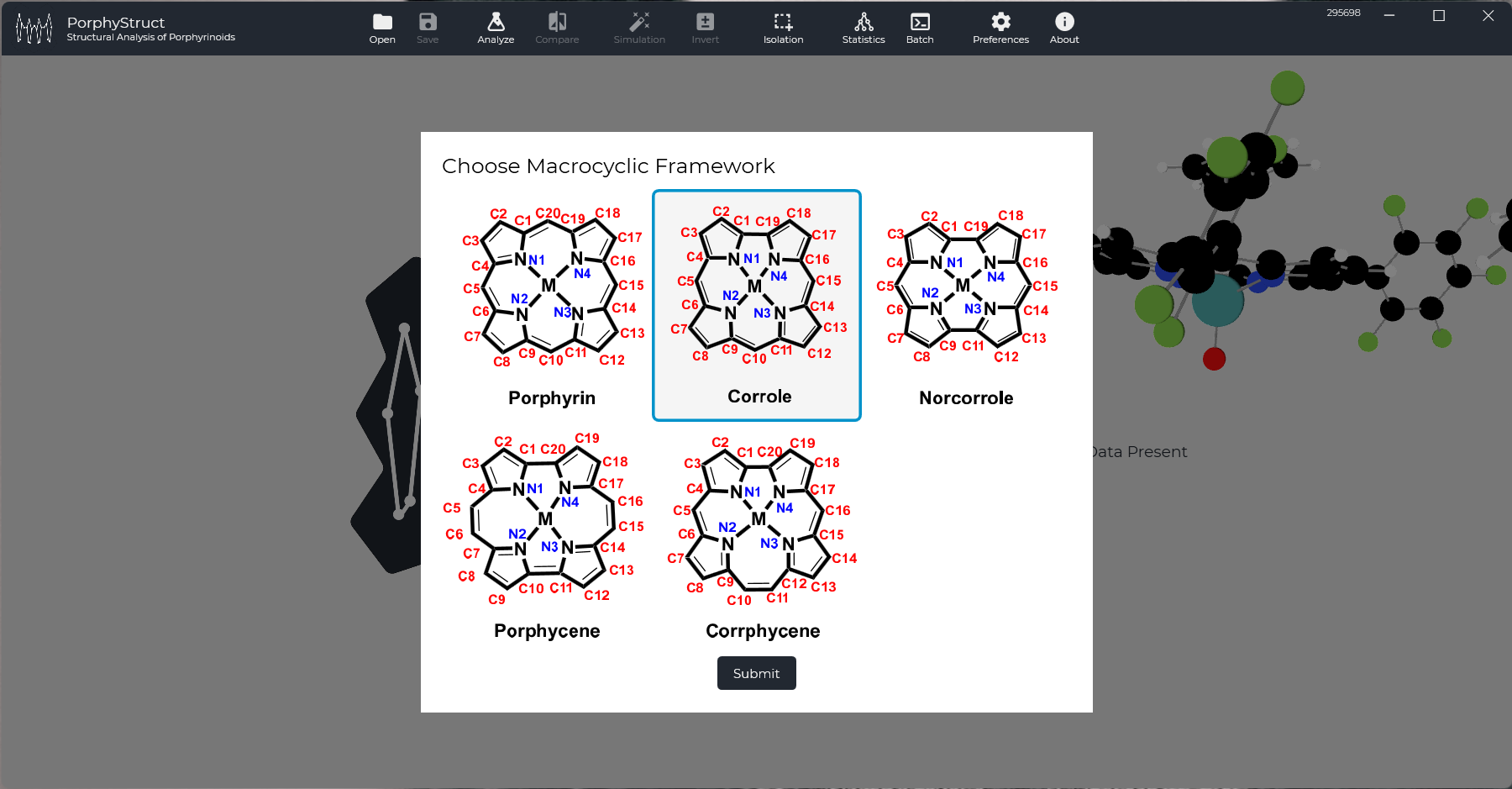
To open a file drag a file onto the PorphyStruct logo in the big bottom left region or click "Open" in the to toolbar. Once you do, a dialog will open and ask for the Macrocyclic Framework. You will also notice, that the Molecular 3D Representation has loaded. (In the picture CCDC File YEBTIJ is used.
Choose your Macrocyle Type and click "Submit". (Please Note: In Version 2.0.0 or higher, this step is automated by PorphyStruct and will not be neccessary)
Generate Analysis
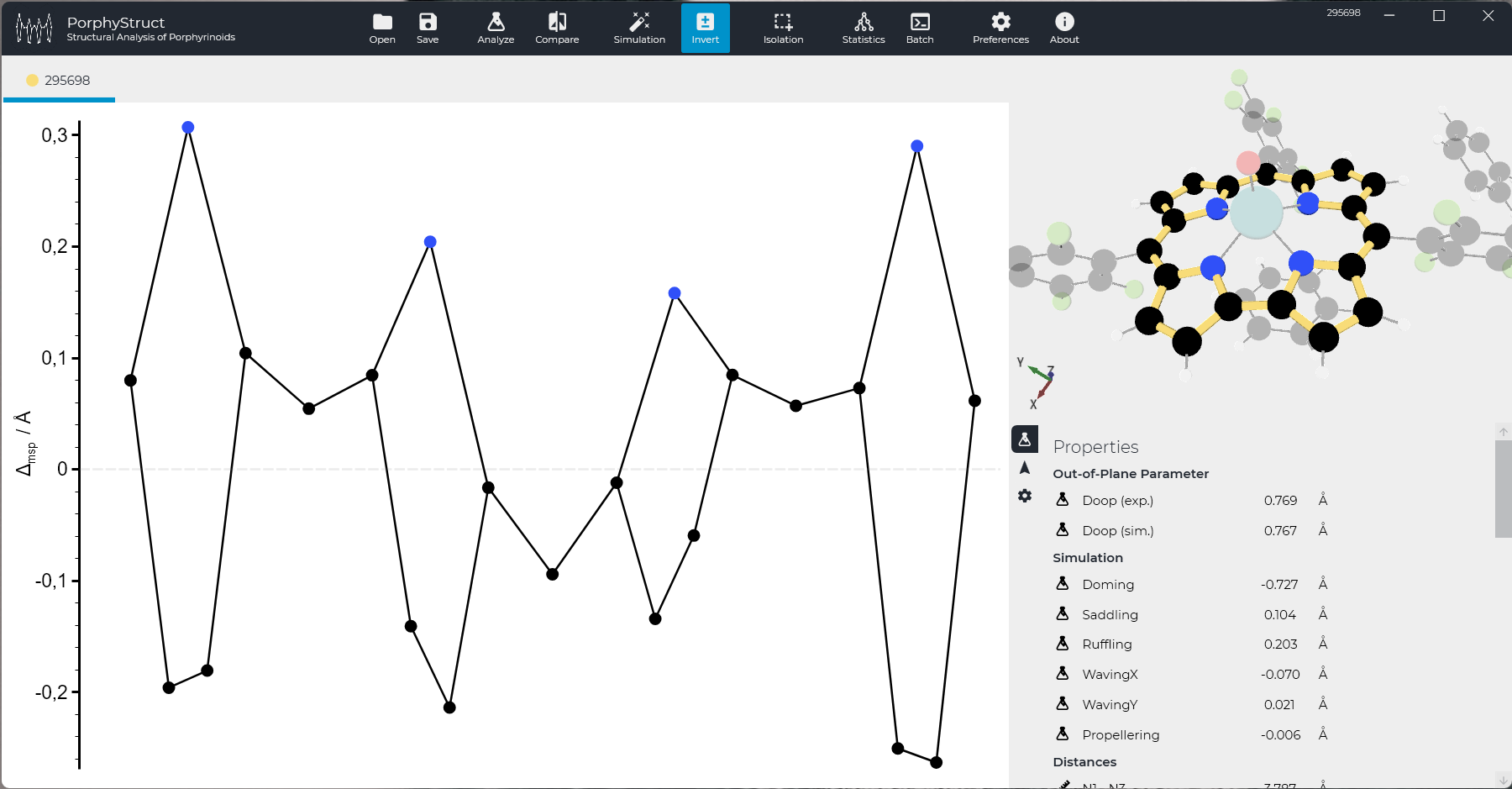
To get what you are here for - the quantitative analysis of the out-of-plane distortions - there are only two clicks left. Click "Analyze" in the Toolbox, a dialog will open. For now click "Minimal Basis" and the analysis will show up. The Macrocyclic Framework is highlighted in a random color (blue in this case) and the displacement diagram is visible. On the right side, all calculated properties will be presented.